Community Network Analysis
Naia Morueta-Holme
Download the PDF of the presentation
The R Script associated with this page is available here. Download this file and open it (or copy-paste into a new script) with RStudio so you can follow along.
1 Setup
library(netassoc)1.1 Generate some random ‘observed’ data
set.seed(1)
# Number of m species
nsp <- 10
# Number of n sites
nsi <- 50
# Observed m x n community matrix (abundance or presence/absence)
m_obs <- floor(matrix(rpois(nsp*nsi,lambda=5),ncol=nsi,nrow=nsp))
# "Force" some species associations to the observed community matrix
m_obs[1,1:(nsi/2)] <- rpois(n=nsi/2,lambda=20)
m_obs[2,1:(nsi/2)] <- rpois(n=nsi/2,lambda=20)1.2 What is the null expectation?
# Null expected m x n community matrix (abundance or presence/absence)
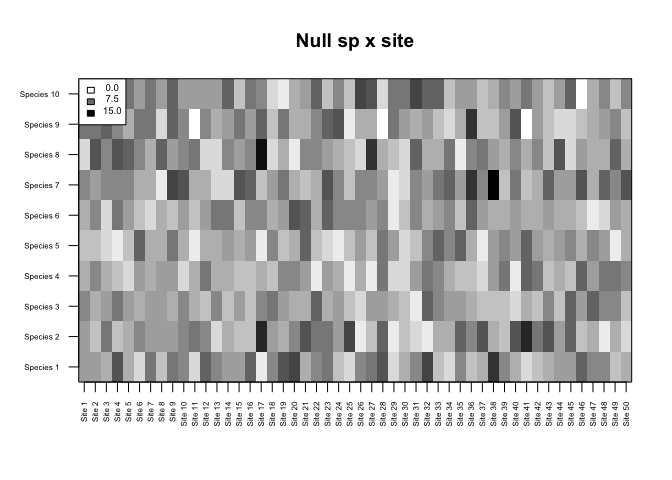
m_nul <- floor(matrix(rpois(nsp*nsi,lambda=5),ncol=nsi,nrow=nsp))Note that here we are simply resampling the observed data preserving row and column sums, which is NOT recommended. Instead, we should use our expected null model of community assembly.
1.3 Infer the species association network
# What species co-occurrence patterns are more or less likely than expected under the null model?
n <- make_netassoc_network(m_obs, m_nul,
method="partial_correlation",
args=list(method="shrinkage"), # for alternative estimators see ?partial_correlation
p.method='fdr',
numnulls=100,
plot=TRUE,
alpha=0.05)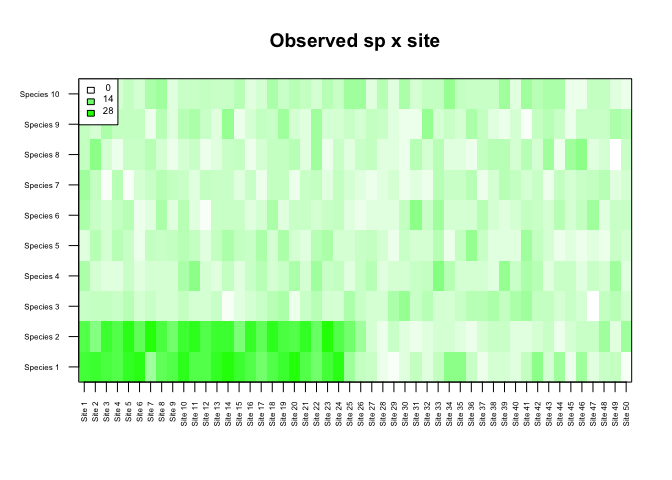
## Calculating observed co-occurrence scores...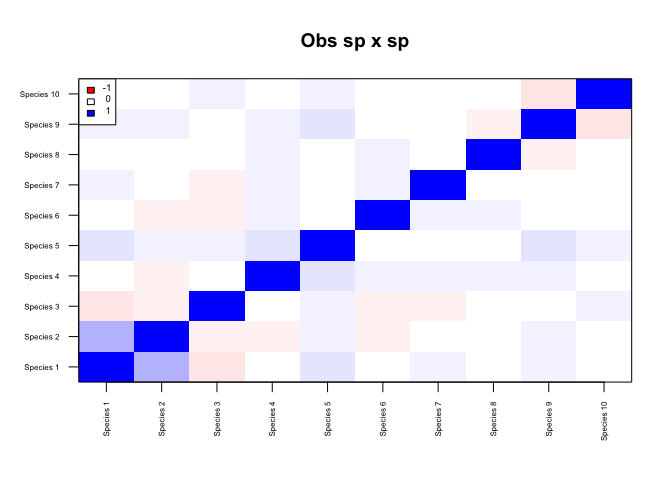
## Generating null replicate 1...
## Generating null replicate 2...
## Generating null replicate 3...
## Generating null replicate 4...
## Generating null replicate 5...
## Generating null replicate 6...
## Generating null replicate 7...
## Generating null replicate 8...
## Generating null replicate 9...
## Generating null replicate 10...
## Generating null replicate 11...
## Generating null replicate 12...
## Generating null replicate 13...
## Generating null replicate 14...
## Generating null replicate 15...
## Generating null replicate 16...
## Generating null replicate 17...
## Generating null replicate 18...
## Generating null replicate 19...
## Generating null replicate 20...
## Generating null replicate 21...
## Generating null replicate 22...
## Generating null replicate 23...
## Generating null replicate 24...
## Generating null replicate 25...
## Generating null replicate 26...
## Generating null replicate 27...
## Generating null replicate 28...
## Generating null replicate 29...
## Generating null replicate 30...
## Generating null replicate 31...
## Generating null replicate 32...
## Generating null replicate 33...
## Generating null replicate 34...
## Generating null replicate 35...
## Generating null replicate 36...
## Generating null replicate 37...
## Generating null replicate 38...
## Generating null replicate 39...
## Generating null replicate 40...
## Generating null replicate 41...
## Generating null replicate 42...
## Generating null replicate 43...
## Generating null replicate 44...
## Generating null replicate 45...
## Generating null replicate 46...
## Generating null replicate 47...
## Generating null replicate 48...
## Generating null replicate 49...
## Generating null replicate 50...
## Generating null replicate 51...
## Generating null replicate 52...
## Generating null replicate 53...
## Generating null replicate 54...
## Generating null replicate 55...
## Generating null replicate 56...
## Generating null replicate 57...
## Generating null replicate 58...
## Generating null replicate 59...
## Generating null replicate 60...
## Generating null replicate 61...
## Generating null replicate 62...
## Generating null replicate 63...
## Generating null replicate 64...
## Generating null replicate 65...
## Generating null replicate 66...
## Generating null replicate 67...
## Generating null replicate 68...
## Generating null replicate 69...
## Generating null replicate 70...
## Generating null replicate 71...
## Generating null replicate 72...
## Generating null replicate 73...
## Generating null replicate 74...
## Generating null replicate 75...
## Generating null replicate 76...
## Generating null replicate 77...
## Generating null replicate 78...
## Generating null replicate 79...
## Generating null replicate 80...
## Generating null replicate 81...
## Generating null replicate 82...
## Generating null replicate 83...
## Generating null replicate 84...
## Generating null replicate 85...
## Generating null replicate 86...
## Generating null replicate 87...
## Generating null replicate 88...
## Generating null replicate 89...
## Generating null replicate 90...
## Generating null replicate 91...
## Generating null replicate 92...
## Generating null replicate 93...
## Generating null replicate 94...
## Generating null replicate 95...
## Generating null replicate 96...
## Generating null replicate 97...
## Generating null replicate 98...
## Generating null replicate 99...
## Generating null replicate 100...
## Calculating standardized effect sizes...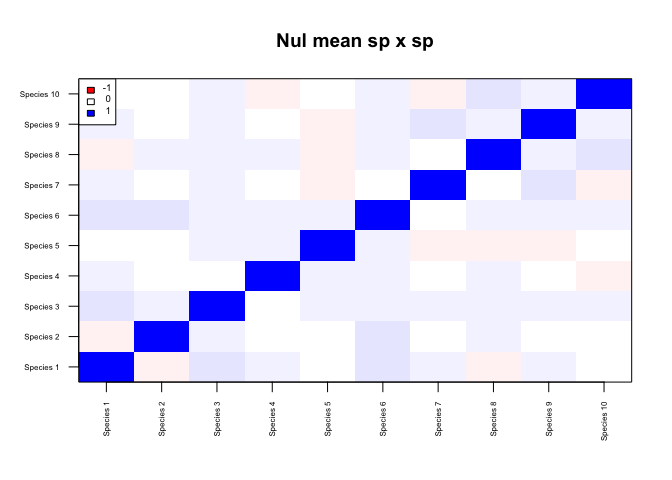
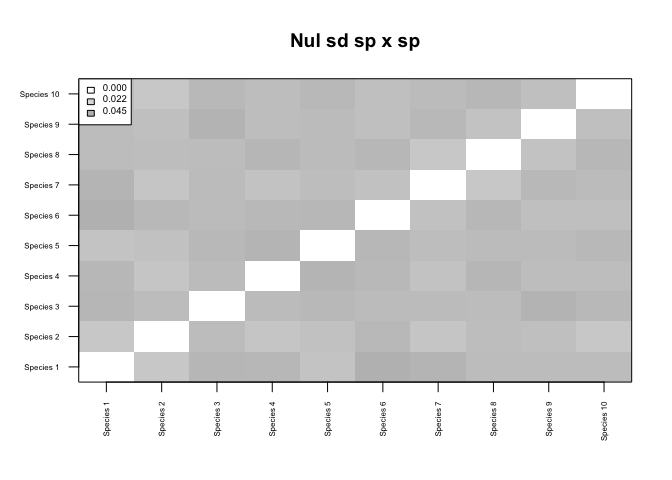
## Adjusting p-values for multiple comparisons...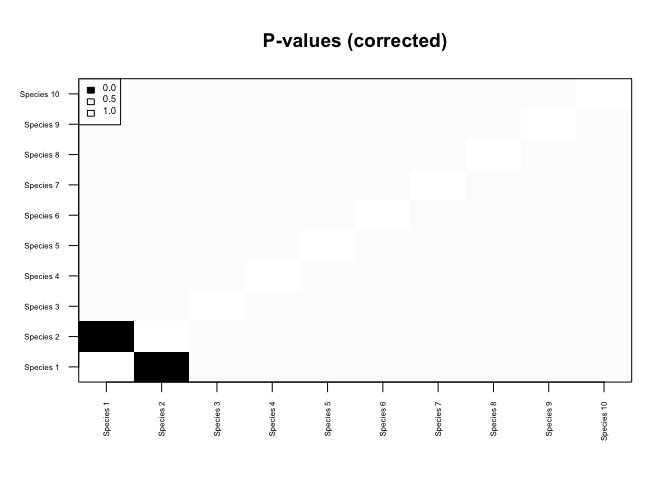
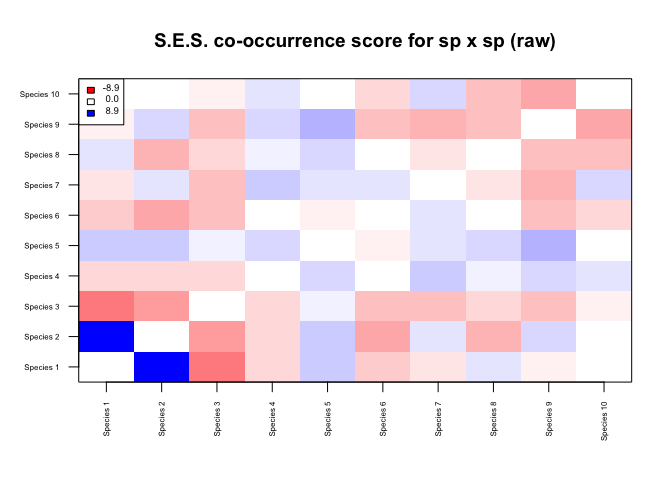
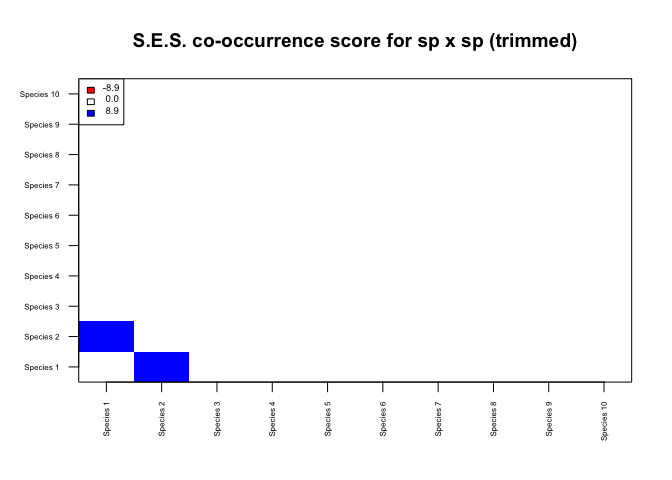
## Building network...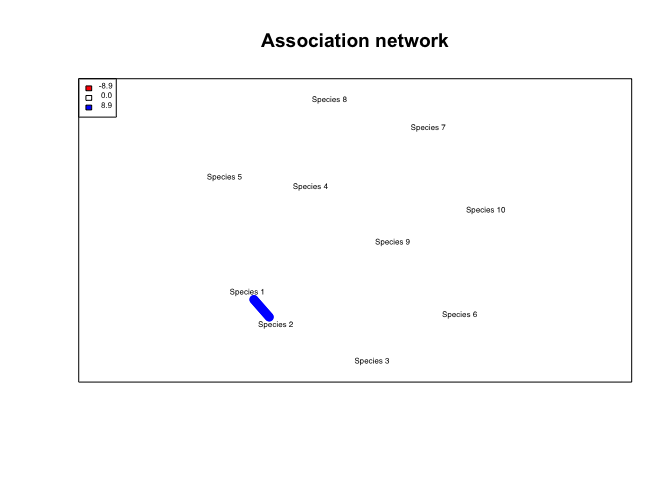
n$network_all## IGRAPH DNW- 10 2 --
## + attr: name (v/c), weight (e/n)
## + edges (vertex names):
## [1] Species 1->Species 2 Species 2->Species 1